Overview
CellCognition Explorer is an open-source image processing tool for the analysis of cellular phenotypes in microscopy. CellCognition Explorer enables phenotype classification by supervised machine learning. To detect rare phenotypes, outlier morphologies can be automatically found by novelty detection methods. A key feature of CellCognition Explorer is an improved classifier training procedure based on automated pre-processing of the full data set into cell gallery images, which can be automatically sorted based on phenotype similarity for efficient iterative classifier training.
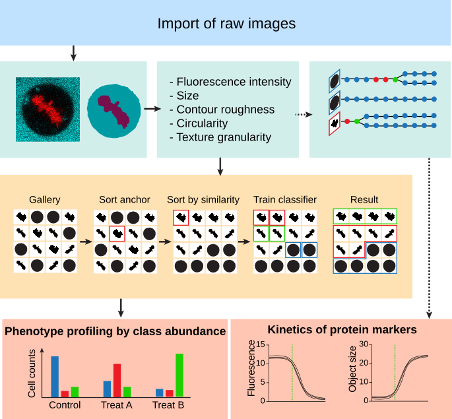
CellCogniton Explorer Software
CellCognition Explorer is released under the GPLv3 and runs on Mac OS X or Windows. The main CellCognition Explorer program provides an integrated solution for image processing, feature extraction, classification by supervised machine learning or novelty detection. In addition, a separate Deep Learning Module program enables to calculate statistical features of cells by deep learning methods. The novelty detection and deep learning methods of CellCognition Explorer have been described in (Sommer et al., 2017).
CellCognition Explorer has been optimized for efficient processing of medium-scale microscopy data. The software is part of the CellCognition project, which provides additional software, e.g., the CeCog Analyzer (Held et al., 2010) for high-throughput by batch-processing with computer cluster support.
Novelty Detection:
As an alternative to manual annotation of phenotype classes, novelty detection methods can be applied to detect outlier phenotypes. This is particularly powerful in case expected phenotypes are very rare. For details, see (Sommer et al., 2017).Deep Learning features
As an alternative to the standard feature set calculated by the main CellCognition Explorer program, the Deep Learning Module calculates features directly based on the pixels of original image data. The Deep Learning Module requires specific hardware for efficient processing. For details, see (Sommer et al., 2017).References
Held, M., Schmitz, M.H., Fischer, B., Walter, T., Neumann, B., Olma, M.H., Peter, M., Ellenberg, J., and Gerlich, D.W. (2010). CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat Methods 7, 747-754.
Sommer, C., Held, M., Fischer, B., Huber, W., and Gerlich, D.W. (2013). CellH5: a format for data exchange in high-content screening. Bioinformatics 29, 1580-1582.
Sommer, C., Hoefler, R., Samwer, M., and Gerlich, D.W. (2017). A deep learning and novelty detection framework for rapid phenotyping in high-content screening. Mol Biol Cell 28, 3428-3436.
Download
CellCognition Explorer has been implemented in Python 3. The program is released under the GPLv3. Binary installers for Microsoft Windows and MacOS X are available here:
CellCognition Explorer main program
- Windows 7/8: CellCognitionExplorer-1.0.2_x86_64.exe
- Max OS-X: CellCognitionExplorer-1.0.2_x86_64.dmg
CellCognition Explorer Deep Learning module
- Windows 7/8: CellCognitionExplorer_v1.0_docker_win7-8_bundle_setup.exe
- Windows 10 : CellCognitionExplorer_v1.0_docker_win10_bundle_setup.exe
CellCognition Deep Learning Module as docker image
The Deep Learning Module is available as CPU-accelerated docker image and available from docker-hub including demo data.
Source code
The program is released as open source software under the terms and conditions of the GPL v3. The source code and bug tracker are hosted on github.
Source installation of the Deep Learning Module
Dependencies
CellCognition Explorer is written in Python 2.7. For Windows users we recommend the Python 2.7 distribution WinPython. Most of the dependencies are included.
Theano
Theano is the main deep learning library we use. Please, follow the general installation instructions for Theano on Windows or Mac OS-X/Linux.
lasagne
Lasagne is a convenient high-level interface to Theano for building deep artificial neural networks.
pip install -r https://raw.githubusercontent.com/Lasagne/Lasagne/master/requirements.txt
pip install https://github.com/Lasagne/Lasagne/archive/master.zip
nolearn
nolearn builds upon lasagne and offers special neural network classes and custom layers.
pip install -r https://github.com/dnouri/nolearn/tree/0.6.0/requirements.txt
pip install nolearn
cellH5
cellH5 is high-content screening data format, which serves as input for CellCognition Explorer and as exchange format to other software such as CellProfiler and Fiji.
pip install git+https://github.com/Cellh5/cellh5@master
CellCognition Explorer - Deep Learning Module
Download cellcognition_explorer_cedl-master.zip and unzip the source code of version 1.0.0. For future releases, please, refer to the code repository on github.
Usage:
General help page
$ python cedl.py --help
Training parameters
$ python cedl.py train --help
Encoding parameters
$ python cedl.py encode --help
Documentation
The novelty detection and deep learning methods of CellCognition Explorer, along with applications to high-throughput RNAi screening data have been described in:Sommer, C., Hoefler, R., Samwer, M., and Gerlich, D.W. (2017). A deep learning and novelty detection framework for rapid phenotyping in high-content screening. Mol Biol Cell 28, 3428-3436.
A detailed software description is available here:
CellCognition Explorer Deep Learning Module
Demo data
Reference data sets representing a broad spectrum of cell morphology phenotypes based on a chromatin marker, are available below. These data have been published in:
Sommer, C., Hoefler, R., Samwer, M., and Gerlich, D.W. (2017). A deep learning and novelty detection framework for rapid phenotyping in high-content screening. Mol Biol Cell 28, 3428-3436.
Data set 1
One can use CellCognition Explorer GUI with ready-made demo data based on 239 morphology feature descriptors and various novelty detection algorithms.
Data set 2
10,000 representative images of HeLa cells expressing histone 2B fused to mCherry (H2B-mCherry) were randomly subsampled from a high-throughput microscopy experiment, as described in Figure 2 in Sommer, Hoefler et al., MBoC, 2017.
- manual_10k_annotated_images.h5 - hdf5 file containing autoencoder features, morphology features, novelty detection prediction, support vector machine prediction and meta data.
- manual_10k_annotated_images.zip - the images as pngs
Data set 3
The Deep Learning Module can be tested with these demo data, which are also included in the software bundle and automatically loaded in the Deep Learning GUI.
Contact
The CellCognition Explorer main software has been developed in the Gerlich laboratory at IMBA, Vienna BioCenter, by Rudolf Hoefler (main CellCognition Explorer software) and Christoph Sommer (Deep Learning Module).
Contact:
- Rudolf Hoefler, Email: rudolf.hoefler@imba.oeaw.ac.at
- Christoph Sommer, Email: christoph.sommer@ist.ac.at
- Daniel Gerlich, Email: daniel.gerlich@imba.oeaw.ac.at