Deep Learning Module
Overview
The Deep Learning Extension enables to learn numerical feature descriptors of cell-objects directly from image pixels using deep convolutional autoencoders.
Prerequisites
The bundled installer contains a demo data set. Starting the Deep Learning Module will automatically load this data set for convenience. If you want to use it on your own data, please, see the list of prerequisites below:
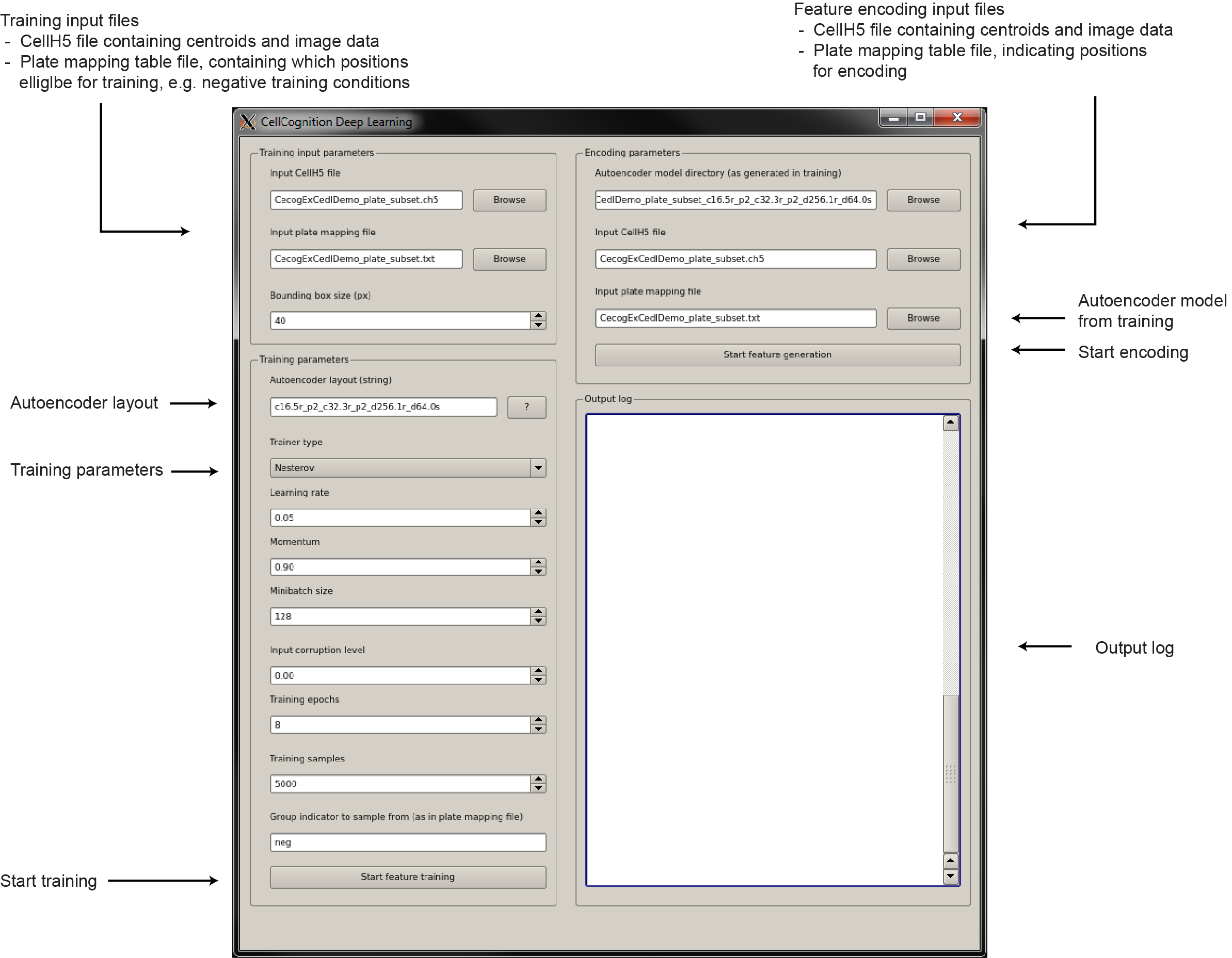
- Cellh5 file containing image data and controids of cell objects. This file can contain multiple positions (perturbation conditions). You can generate this with the CellCognition Analyst software or with CellProfiler using the exportToCellh5 module.
- Approximate size of cell-objects in pixels, i.e. the typical width and height of an enclosing rectangle
- A position mapping text file containing a table, where each row maps the position ID as found in the CellH5 file (e.g. C03) to a perturbation condition (neg. control)
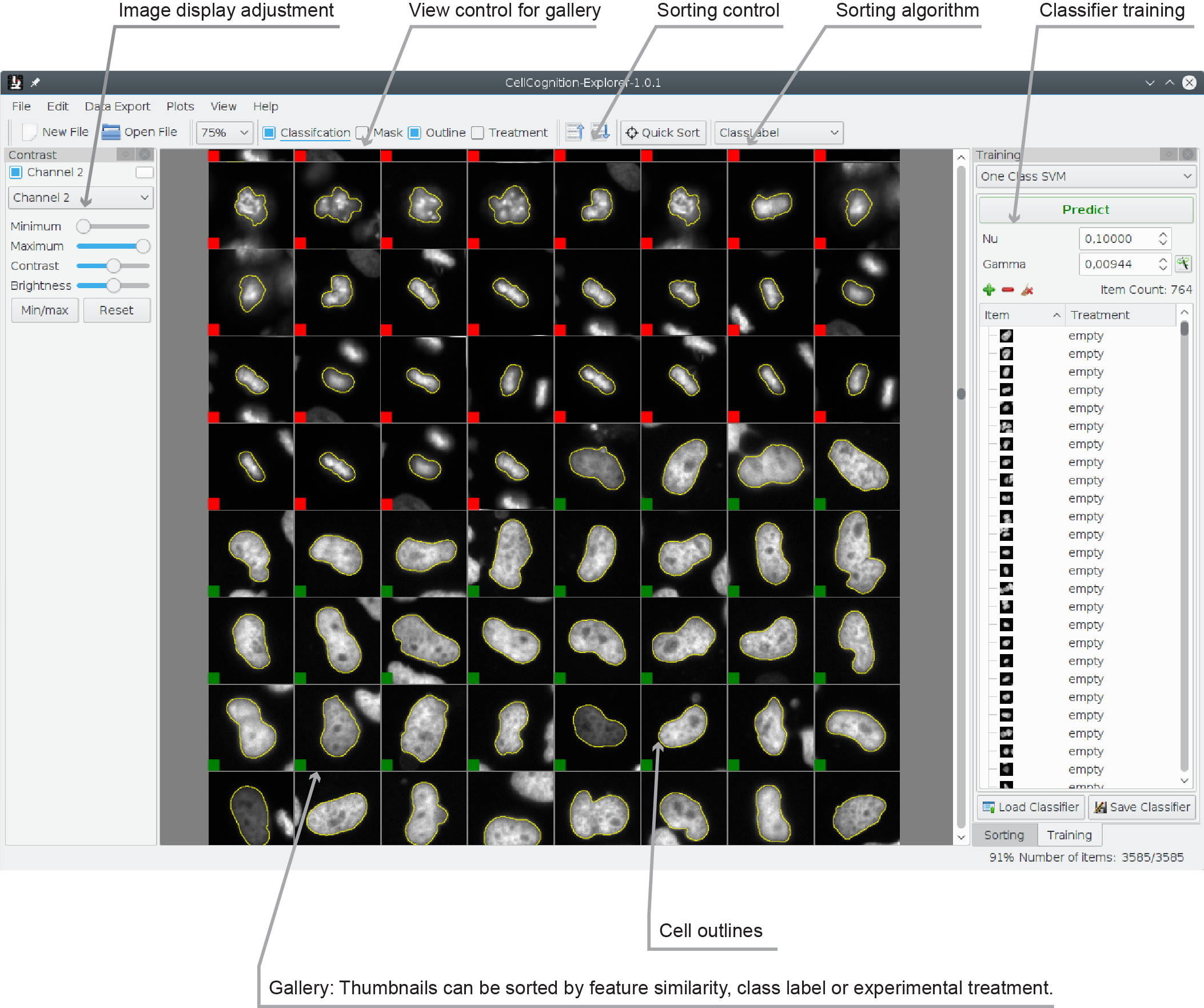
The graphical user interface
The graphical user interface is quite simple.
How to choose good training parameters?
The optimization strategy
Training an autoencoder is a rather complex multi-parametric optimization problem. The optimization algorithm works iteratively. Each training round is called an epoch. In each epoch, the current status of the autoencoder is evaluated and the algorithm proposes how to change each internal parameter of the autoencoder to achieve a better performance in the next round. The intensity of this proposed change of each internal parameters can be chosen. However, it is not guarenteed, that the autoencoder acutally improves after an epoch on held out data.
The tuning parameters
- learning rate: gives the strength of each internal parameter adjustment in each epoch. Choosing too high values lead to oscillation behaviour in the optimization. Too low values, lead to slow convergence and many epochs will be needed. A good range of values for the learning rate are from 0.01 to 0.1 for both the AdaGrad and Nesterov optimization algorithm.
- momentum: specifies the fraction of the last parameter adjustments, from the last epoch, that should be incorporated into the current estimation of the parameter adjustments. This parameter can prevent oscillation and helps to improve convergence (less training epochs). Good default values are from 0.5 to 0.9. Note, this parameter is only available for the Nesterov training procedure
- Minibatch size:In each epoch all available training images are chunked into batches of a certain size. The evaluation then is averaged over all images in each batch. Choosing too big batche sizes will average out finer structures found in the image data. Too small values will lead to a significant increase of computation time. Good values range from 32 to 256.
- input corruption level:An autoencoder aims to reconstruct its input image as good as possible. It has been shown that the convergence can be accelerated when the autoencoder reconstructs from an corrupted image. The corruption level gives the strength of the corruption as a fraction of 0 to 1. Good choices range from 0 to 0.3.
- Training epochs: sets the number of epochs used in the iterative training. The more the better, however, in the beginning of the training the relative improvement after each epoch is stronger than later. When choosing Training epochs too big, almost no substantial increase in performance will be made in later epochs. Good choices start from 64 to 1000, depending on the image complexity.
Visualizing results
After cell-object features have been learned with the Deep Learning Module, start the CellCognition Explorer main graphical user interface and open the generated file (.hdf). It should look similar to this: